All organisms release DNA into the environment. Sampling and analysing this environmental DNA (eDNA) promises an elegant solution to detect…
Liesel Morgan
Land snails often exhibit high levels of endemism and are vulnerable to habitat degradation. As such, they are often targeted during Biologic’s short-range endemic (SRE) surveys. Land snails are generally active during wet weather, and they can also be found buried in leaf litter or beneath logs where they aestivate (a form of hibernation) until conditions are favorable. In the Pilbara, one of the snail families we target is the Camaenidae, snails of the genera Rhagada and Quistrachia. Snail species identification usually requires dissection of body parts, specialist skills that necessitate the euthanasia of the snail. Shell morphology is also used, however a number of studies have shown that shell morphology may vary within a species and cannot be relied upon for species identification1.. Our molecular systematics team at Biologic uses genetic techniques (DNA barcoding) to identify individuals. This also requires snail tissue being taken, which again entails euthanasia.
A common issue with identifying snails (either by dissection of body parts or removal of tissue for molecular analysis) is their ability to retract into their shells and seal themselves in. This then leads to the risk that they expire within their shells and proceed to decompose. This decomposition damages soft tissue and DNA rendering them unidentifiable. Having to euthanise an animal that is potentially ecologically significant also raises ethical questions about removing a specimen from the environment. Recently there have been advances in the passive sampling methods for mygalomorph (trapdoor) spiders to conserve these rare and potentially significant species, so we investigated whether it was possible to do this for land snails.
After some research into techniques used for other gastropods, we conducted a trial using cotton swabs on live Rhagada to see if a mucus sample would be sufficient as a DNA sample. Individuals were encouraged out of their shells and onto sterile swabs to ‘walk’ around and leave a trail of mucus (see adorable images). These swabs were then treated like a regular sub-sample of tissue and placed into buffer for DNA extraction, amplification and sequencing. From eleven individuals, we were able to get nine sequences of Rhagada snails, a fantastic success rate that is comparable to current subsampling methods. We plan to test this non-destructive method under field conditions in the future, where individuals can be released directly back into their habitat. We hope that we can introduce this method as a standard way to passively sample snails and return them to their habitats to live out their slow-moving lives.
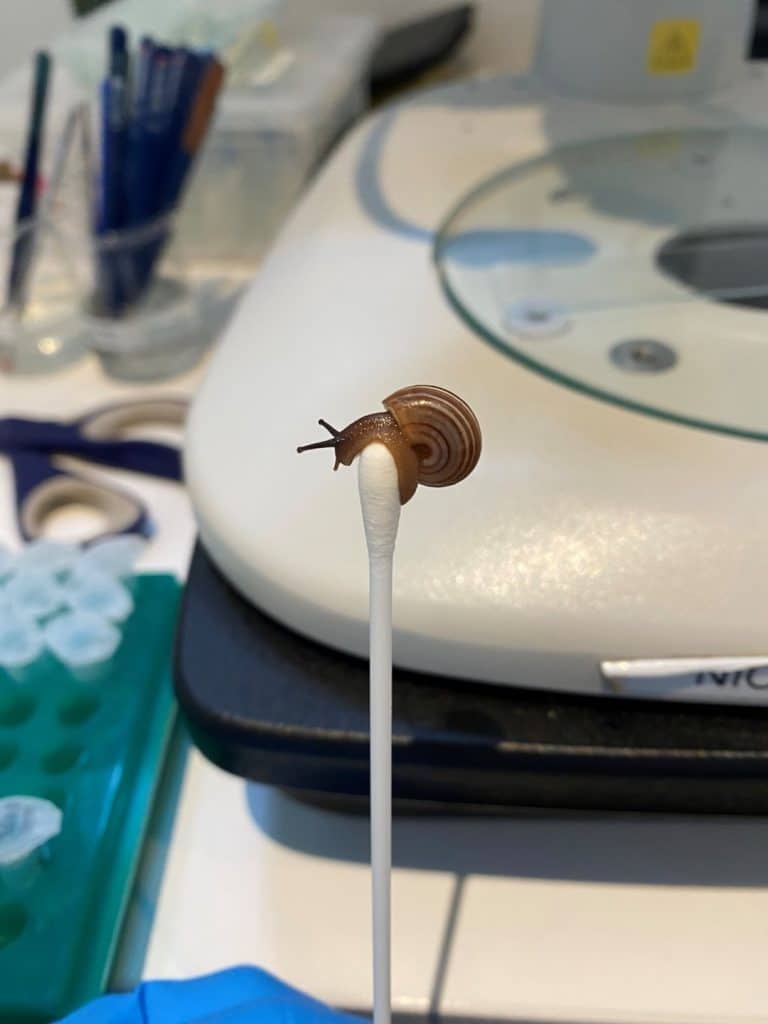
Figure 1: Rhagada species walking onto a sterile swab.
1Hamilton, Z. R. (2021). Repeated evolution of an undescribed morphotype of Rhagada (Gastropoda: Camaenidae) from the inland Pilbara, Western Australia. Invertebrate Systematics, 35(2), 203-215.
1Hamilton, Z. R., & Johnson, M. S. (2015). Hybridization between genetically and morphologically divergent forms of Rhagada (Gastropoda: Camaenidae) snails at a zone of secondary contact. Biological Journal of the Linnean Society, 114(2), 348-362. doi:10.1111/bij.12410
1Johnson, M. S., Hamilton, Z. R., Kendrick, P. G., & Teale, R. (2012). Endemic evolutionary radiation of Rhagada land snails (Pulmonata: Camaenidae) in a continental archipelago in northern Western Australia. Biological Journal of the Linnean Society, 106, 316-327.
1Johnson, M. S., & Stankowski, S. (2018). Extreme morphological diversity in a single species of Rhagada (Gastropoda: Camaenidae)inn the Dampier Archipelago, Western Australia: review of evidence, revised taxonomy and changed perspective. Journal of Molluscan Studies, 84, 337-344.
1Johnson, M. S., Stankowski, S., Kendrick, P. G., Hamilton, Z. R., & Teale, R. J. (2016). Diversity, complementary distributions and taxonomy of Rhagada land snails (Gastropoda : Camaenidae) on the Burrup Peninsula, Western Australia. Invertebrate Systematics, 30, 323-334. doi:10.1071/IS15046



